pET-39b(+) Vector
Anuncio

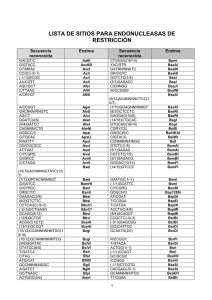
TB230 12/98 pET-39b(+) Vector The pET-39b(+) vector (Cat. No. 70090-3) is designed for expression of DsbA fusion proteins. Unique sites are shown on the circle map. Note that the sequence is numbered by the pBR322 convention, so the T7 expression region is reversed on the circle map. The cloning/expression region of the coding strand transcribed by T7 RNA polymerase is shown below. The f1 origin is oriented so that infection with helper phage will produce virions containing single stranded DNA that corresponds to the coding strand. Therefore, single stranded sequencing should be performed using the T7 terminator primer (Cat. No. 69337-3). Avr II(131) Xho I(174) Eag I(182) Not I(182) Hind III(189) Sal I(195) Sac I(206) EcoR I(208) BamH I(214) Nco I(227) Srf I(246) BseR I(250) Kpn I(271) Nsp V(301) Sca I(341) Sac II(365) SpeI(396) pET-39b(+) sequence landmarks 1103-1119 1102 408-1031 369-386 282-326 174-250 150-173 26-72 1510-2589 4023 4732-5544 5640-6094 Dra III(5864) Kan (47 32 -5 5 Pvu I(5163) Sgf I(5163) Nru I(4820) ) 44 640-6094) n (5 rigi o f1 Afl II(418) Pst I(552) BspM I(636) Ds bA Asc I(917) PinA I(928) (4 0 ) 31 10 8- T7 promoter T7 transcription start DsbA•Tag™ coding seq. His•Tag® coding sequence S•Tag™ coding sequence Multiple cloning sites (Srf I - Xho I) His•Tag coding sequence T7 terminator lacI coding sequence pBR322 origin Kan coding sequence f1 origin Nde I(1030) Xba I(1068) SgrA I(1179) Sph I(1335) pET-39b(+) (6106 bp) Eco57 I(4509) o ri (40 BssS I(4134) 23 ) 9) BspLU11 I(3961) lacI (1 5 1 0-2 58 AlwN I(4377) Mlu I(1860) Bcl I(1874) Apa I(2071) Sap I(3845) Bst1107 I(3732) Tth111 I(3706) PshA I(2705) Bgl I(2924) Fsp I(2942) Psp5 II(2967) T7 promoter pET upstream primer #69214-3 lac operator rbs Xba I ATGCGTCCGGCGTAGAGGATCGAGATCGATCTCGATCCCGCGAAATTAATACGACTCACTATAGGGGAATTGTGAGCGGATAACAATTCCCCTCTAGAAATAATTTTGTTTAACTTTAAGAAGGAGATATACAT S-DsbA•Tag primer #70177-3 DsbA ATGAAAAAGATTTGGCTGGCGCTGGCTGGTTTAGTTTTAGCGTTTAGCGCATCGGCGGCG...522bp...CAGCAGTATGCTGATACAGTGAAATACTTAAGCGAGAAAAAAGGATCAACTAGTGGTTCTGGT ˜È„ÒÅÒÅÌÒ◊ÒÅÏÍÅÍÅÅ...174aa...ŒŒÁÅÎÊ◊ÁÒ͉ÌÍÊÍÌÍÌ signal peptidase His•Tag S•Tag Sca I S•Tag primer #69945-3 Nsp V Kpn I CATCACCATCACCATCACTCCGCGGGTCTGGTGCCACGCGGTAGTACTGCAATTGGTATGAAAGAAACCGCTGCTGCTAAATTCGAACGCCAGCACATGGACAGCCCAGATCTGGGTACC ÓÓÓÓÓÓÍÅÌÒ◊∏ÂÌÍÊÅÈ̘‰ÊÅÅÅωŒӘÎÍ∏ÎÒÌÊ Srf I thrombin BseR I Nco I BamH I EcoR I Sac I Sal I Hind III Eag I Not I Xho I His•Tag GATGACGACGACAAGAGCCCGGGCTTCTCCTCAACCATGGCGATATCGGATCCGAATTCGAGCTCCGTCGACAAGCTTGCGGCCGCACTCGAGCACCACCACCACCACCACCACCACTAA ÎÎÎÎÍ∏ÌÏÍÍʘÅÈÍÎ∏ˆÍÍÍ◊ÎÒÅÅÅÒ‰ÓÓÓÓÓÓÓÓEnd enterokinase Avr II Bpu1102 I T7 terminator TTGATTAATACCTAGGCTGCTAAACAAAGCCCGAAAGGAAGCTGAGTTGGCTGCTGCCACCGCTGAGCAATAACTAGCATAACCCCTTGGGGCCTCTAAACGGGTCTTGAGGGGTTTTTTG T7 terminator primer #69337-3 pET-39b(+) cloning/expression regions Novagen • FAX 608-238-1388 • E-MAIL [email protected] pET-39b(+) Restriction Sites Enzyme AccI AciI AflII AflIII AluI AlwI Alw26I # Sites 2 84 1 2 27 13 6 AlwNI ApaI ApaLI ApoI 1 1 3 7 AscI AvaI AvaII AvrII BamHI BanI 1 3 5 1 1 10 BanII 7 BbsI BbvI BcgI BcgI’ BclI BfaI 5 27 4 4 1 8 BglI BglII BpmI Bpu10I Bpu1102I BsaAI BsaBI BsaHI BsaJI BsaWI 1 2 5 2 2 2 3 5 15 8 BseRI BsgI BsiEI BsiHKAI 1 3 5 7 BslI BsmI BsmBI BsmFI Bsp1286I BspEI BspLU11I BspMI BsrI BsrBI BsrDI BsrFI 27 2 3 4 13 2 1 1 21 4 2 8 BssHII BssSI Bst1107I BstEII BstXI BstYI 2 1 1 2 4 9 Cac8I ClaI CviJI DdeI DpnI DraIII DrdI 45 2 93 12 24 1 3 Locations 196 3731 418 1860 1557 5179 4377 2071 1840 208 5666 917 174 2412 131 214 267 1780 206 4818 710 3961 1962 3775 303 5677 244 2788 352 2499 251 5939 2006 2088 4275 2135 5035 2876 2475 4776 2967 1317 5901 2071 2345 2719 3079 2720 2686 3572 3538 132 4763 397 6015 1069 2187 2821 2 4167 250 1711 185 175 3779 928 4314 2179 5298 1911 2645 206 4279 3121 3877 1360 5047 2475 1321 5124 3602 2862 2 3961 636 3150 1089 1907 928 2918 917 4134 3732 773 468 214 3153 3894 2273 1170 5117 2271 1137 4854 5864 3654 4069 3246 1203 2755 1258 2152 2186 2041 1662 274 4602 4960 1182 2629 1244 210 176 1874 70 4456 2924 274 888 3067 80 3713 1133 1183 760 1698 5180 564 5864 1143 1204 3602 3158 1318 5179 3232 2500 2682 3150 4301 1844 5163 2955 6079 6008 1179 5965 1546 5819 3488 1817 5562 1791 760 4613 2975 1914 1424 5412 2758 2636 TB230 12/98 Enzyme DsaI EaeI EagI EarI Eco47III Eco57I EcoNI EcoO109I EcoRI EcoRII EcoRV FauI Fnu4HI FokI # Sites 4 4 1 4 3 1 2 3 1 11 2 19 48 10 FspI HaeII HaeIII HgaI HhaI HincII HindIII HinfI HpaI HphI KpnI MaeIII MboII MluI MnlI MseI MslI 1 16 23 12 52 4 1 20 2 21 1 18 19 1 26 29 6 MspI MspA1I MunI MwoI NarI NciI NcoI NdeI NgoAIV NlaIII NlaIV NotI NruI NsiI NspI NspV PflMI PinAI PleI 32 13 2 43 4 14 1 1 4 28 23 1 1 2 4 1 3 1 10 PshAI Psp1406I Psp5II PstI PvuI PvuII RcaI RsaI 1 4 1 1 1 5 3 8 SacI SacII SalI SapI Sau3AI Sau96I ScaI ScrFI SfaNI SfcI 1 1 1 1 24 14 1 25 24 5 Locations 227 362 182 1168 182 618 1478 1265 2766 4509 1395 5075 53 1293 208 222 653 444 3320 2942 1906 3506 197 189 509 509 2366 1297 1300 2933 2534 3845 3215 4976 2967 1915 3647 791 3180 4801 3242 5407 Enzyme SgfI SgrAI SmaI SpeI SphI SrfI SspI StyI TaiI TaqI TfiI # Sites 1 1 2 1 1 1 2 3 16 17 10 ThaI TseI Tsp45I 40 27 9 Tsp509I TspRI Tth111I VspI 24 14 1 6 XbaI XcmI XhoI XmnI 1 3 1 4 2366 Locations 5163 1179 246 5037 396 1335 246 5088 5656 57 131 227 488 3936 2539 5074 2841 5130 3011 5302 3515 5393 708 3613 773 3708 2041 5310 2869 6037 3400 1117 2545 2604 5362 2232 2250 861 3519 3706 139 5551 1068 1716 174 592 5552 271 Enzymes that do not cut pET-39b(+): AatII AhdI BsaI BsrGI DraI FseI MscI NheI PacI PmlI RsrII SanDI SexAI SfiI Sse8387I StuI SunI SwaI 1860 1912 3534 2200 332 534 1183 227 1030 1170 182 4820 5013 1335 301 293 928 763 3855 2705 1522 2967 552 5163 494 1258 269 2007 206 365 195 3845 2230 2948 3143 1204 1318 2500 2758 2918 5965 5279 3306 3598 3965 1442 5426 1117 4340 1409 5395 1496 5799 2890 3286 5649 547 4681 341 3767 2460 5556 610 4998 2553 3552 687 949 1102 4226 4417 6083 2292 5807 341 548 Novagen • FAX 608-238-1388 • E-MAIL [email protected] Bsu36I PmeI SnaBI UbaEI