Metabolómica in vivo y ex vivo en el control de la Actividad Farmacológica B. Celda1,2,3, D. Monleón2,3,4, MC. Martínez1,2,3, V. Esteve1,2,3, B. Martínez1,3, E. Piñeiro1, R. Ferrer1 1Dept. Química Física-SCSIE, Universitat de Valencia, 2Laboratorio Imagen Molecular (UCIM) 3CIBER Bioingeniería, Biomateriales y Nanomedicina, ISC III 4Fundación de Investigación del Hospital Clínico Universitario de Valencia
Anuncio

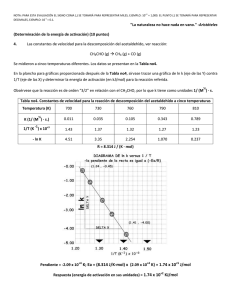
Metabolómica in vivo y ex vivo en el control de la Actividad Farmacológica B. Celda1,2,3, D. Monleón2,3,4, MC. Martínez1,2,3, V. Esteve1,2,3, B. Martínez1,3, E. Piñeiro1, R. Ferrer1 1Dept. Química Física-SCSIE, Universitat de Valencia, 2Laboratorio Imagen Molecular (UCIM) 3CIBER Bioingeniería, Biomateriales y Nanomedicina, ISC III 4Fundación de Investigación del Hospital Clínico Universitario de Valencia (FP6-2002-LIFESCIHEALTH 503094) Esquema Metabolómica y Actividad Farmacológica 1.- Introducción 2.- Precisión diagnóstico Selección Terapia in vivo, ex vivo e in vitro 3.- Sistemas Modelo in vivo, ex vivo e in vitro 4.- Desarrollo Bioinformático BIONÓMICA ADN ARN Genómica Chips ADN SCSIE+UCIM 40.000 genes Conocimiento Limitado Transcriptómica 105 transcripciones Chips ADN SCSIE+UCIM Proteínas Proteómica 109 proteínas MALDI-Tof MALDI-Tof RMN SCSIE Metabolitos Metabolómica 3000 metabolitos Metabonómica estructura conocida (RMN) RMN Alto Campo SCSIE+UCIM B.Celda Conocimiento Adecuado Aplicaciones Moleculares al Diagnóstico, Pronóstico y Selección Terapia Medicina Translacional Metabolómica (in vivo y ex vivo) Genómica (Transcriptómica y Genotipado) Proteómica (Funcional y Estructural) B.Celda INTRODUCCION NMR Applications 1H( MHz) Magnetic Field Intensity (T) Size (cm) 9,6 - 100 0,25 - 3,0 < 100 MRI + 41 - 300 MRS (biomedicine) 1,0 - 7,1 Miniimaging 300 - 400 NMR Microscopy 200 - 600 Type resonance frequency MRI + MRS (clínical) NMR 400 - 900 multidimensional B.Celda Spatial resolution (mm3) Application System d5 human body < 30 d2 small animals 7,1 - 9,4 < 12 # 0,005 mouses 4,7 - 14,1 < 2,5 0,00005 insects, materials, embrions 9,4 - 21 < 40 Å < 30 d2 small animals Laboratorio Imagen Molecular (rabits, mouses…) UCIM (UVEG) proteíns, DNA, tissues ... PERFIL BIOQUÍMICO Espacio Intra y extracelular, Fluidos corporales Metaboloma Fluidos Corporales RMN Metabonoma Metabolómica Plantas Metabolómica Mamíferos Metabolómica humanos: Cerebro Corazón Hígado: Hepatocito Riñón Próstata Cáncer Mama Tejido parenquimal Tejido Médula Líquido Cefalorráquideo Sangre: Plasma Suero Bilis Fluido Seminal Orina Exposición por enfermedad, cambio micro-flora, fármacos, tóxicos ... B.Celda In vivo + ex vivo Metabolism Genetic profile (transcriptomics) Protein expression (Proteomics) Metabolism (Metabolomic) Preceds to tissue physiological changes B.Celda Brain Tissue Metabolite Profile by SV 1H MRS B.Celda Alzheimer NAA mI Co TE = 31 ms Cr NAAp ; mIn NAA/mIp NAA/mI d 2,32 ERM 1 0.9 Cr NAA Global 0.8 RMI 0.7 TE = 136 ms Sensibilidad Cr 0.6 0.5 0.4 Europ.J.Neurology 2004: 11; 89-98 0.3 NAA/Crp 0.2 0.1 0 0 0.2 0.4 0.6 0.8 1 - Espe cificidad (fals os positivos) B.Celda 1 MRSI vs. PET B A NAA Co Cr Lac NA Metastatic Cystic MRSI vs. PET B A 3 1 6 4 2 7 5 C High Grade Glioma Brain tumour Diagnosis MRSI + SV SV location of AII according Cho High level previous 2D TSI TE 272 ms MMA/Cr TR 2s 1.5 T MMB/Cr (FP6-2002-LIFESCIHEALTH 503094) 3 4 Gliomas grading by two TE (31 + 136 ms) SV quantitative analysis previous SV location by 2D TSI high level Choline image Grade 2 http:// www.etumour.net, M.C. Martínez-Bisbal et al. ESMRMB (2005); # 114 and Rev.Neurol. (2002), 34(4); 309 C. Majós et al. AJNR (2004), 25; 1696; http://www.carbon.uab.es/INTERPRET, B.Celda Brain primary tumour vs metastasis MRSI 2D TSI at 272 ms 1.5 T Brain secondary lesion metastasis (MT) (Cho/Cr)MT<(Cho/Cr)GBM Brain brimary tumour (GBM) (FP6-2002-LIFESCIHEALTH 503094) B.Celda http:// www.etumour.net, Brain tumour patient outcome and follow-up by Perfusion MRI and MRS Kaplan-Meier survival curves Cerebral Blood Volume (CBV) predicts more accurately the Low Grade Gliomas (LGG) progression than initial histopathology grading Longer survival o CBV<1.75 Shorter survival o CBV>1.75 M. Law et al. Proc. Intl. Soc. Mag. Reson. Med. 13 (2005); 330 Follow-up of AII by 1H MRS SV at 1.5 T TE 136 ms Cho Cr NAA MRS study number B.Celda Guided Therapy; Radiotherapy, Quimio Tumour tissue Infiltration A Normal tissue B Necrosis T. Laudatio, MC. Martínez-Bisbal et al., NMR Biomedicine, in press (2006) B.Celda Brain tumour radionecrosis vs. recurrence MRSI Transverse 2D MRSI 1.5 T TE 31 ms TR 2 s Cho Tumour Recurrence (TR) (Cho/NAA)TR >(Cho/NAA)RI Co Saggital 2D TSI 1.5 T TE 272 ms TR 2 s Radiation injury (RI) B.Celda http:// www.etumour.net, (FP6-2002-LIFESCIHEALTH 503094) Breast cancer diagnosis (MRS/CSI) 2D PRESS MRSI (CSI) with k-space sampling 1.5 T TE = 270 ms 16 x 16 11:40 min 8 averages Two independent CHESS optimized pulses for water and lipid suppression 2D 1H MRSI study after 5 days no Cho Gd-enhanced areas absence of viable tumour High Cho levels active tumour by histology J. Hu et al. Proc.Intl.Soc.Mag.Reson.Med. 13 (2005); 134 Liver treatment response by 19F MRSI at 3 T of Capacitabine colorectal cancer drug 19F CSI of liver of a patient treated with oral Capecitabine FABL imaging Capecitabine Capecitabine o 5’ deoxy-5’ fluorocytidine o 5’ deoxy-5’ fluorouridine o D-fluor-E-alanine Oral drug 5’ DFUR 5’ DFCR FBAL D.W. Klomp.et al. Proc. Intl. Soc. Mag. Reson. Med. 13 (2005); 128 Ex Vivo 1H MRS Lac Leu Ala PCho mI Germinoma Val ?? Ile ?? Leu Gly Tau?? Cho Leu PCr Cr Glx NAA Ile NAA Asp GBM NAA lip Biopsy study by HR-MAS (ex vivo) HR-MAS PCA T=0ȱCȱ(4ȱCȱinner) 4500ȱHz ca.ȱ20ȱmgȱofȱtissue Cylindricalȱrotor (50ȱPl) (PCAȱca.ȱ2g) D2Oȱfieldȱhomogeneity andȱmobility HR-MAS PCA NMR in Biomedicine, Martínez-Bisbal et al. 2004;17:191-205 B.Celda 129 Brain tumour characterization ex vivo (HR-MAS) Met Ast Men GBM (FP6-2002-LIFESCIHEALTH 503094) B.Celda M.C. Martínez-Bisbal et al. NMR Biomed.(2005), 17; 1 http:// www.etumour.net; D. Monleón et al., ESMRMB (2005) #325 Brain tumour biopsies ex vivo metabolic profile NMR Biomed. 2004;17:191-205 Brain tumour characterization ex vivo (HR-MAS) (FP6-2002-LIFESCIHEALTH 503094) B.Celda M.C. Martínez-Bisbal et al. NMR Biomed.(2005), 17; 1 http:// www.etumour.net; D. Monleón et al., ESMRMB (2005) #325 Fle 158 Metastasis In vivo ERM 30 ms a 1,5 T ex vivo HR-MAS a 11,5 T in vivo ERM 30 ms a 1,5 T Fle 153 Ganglioglioma ex vivo HR-MAS a 11,5 T Perfil Metabólico ex vivo Biopsias Hígado Glucosa E/LipI Lip I/LipII Alanina/Lisina p < 0,001 p = 0,01 p = 0,026 NMR Biomed. 2006;19:90-100 Breast cancer diagnosis and invasion High-Resolution 1H NMR malignant bening 1H NMR spectra at 8.5 T, 37 C of breast fine needle aspiration biopsy Accuracy Accuracy benign vs malignant 96% (Cho/Cr) lymph node vascular invasion 95% 94% 1H NMR spectra at 8.5 T, 37 C of breast ductal carcinoma in situ (DCIS) fine needle biopsy C.E. Mountford et al. Chem. Rev. (2004), 104; 3677 C.E. Mountford et al. Br. J. Surg. (2001), 88; 1234 C. Lean et al., J. Women´s Imaging (2000), 2; 19 Breast cancer grading and lymph node status (HR-MAS) 1H HR-MAS CPMGPR aliphatic region of a breast tumour tissue at 14 T • Breast cancer grading for invasive ductal carcinoma IDC IDC I IDC II IDC III Sensitivity 100% 84% 81% Specificity 100% 83% 86% B) Breast cancer lymph node status prediction by HR-MAS Actual negative Sensitivity 92% Specifity 97% Actual Positive 97% 92% B. Sitter et al. NMR in Biomedicine 2002; 15: 327 T.F. Bathen et al., Proc. Intl. Soc. Reson. Med., 13 (2005), 130 L.R. Jensen et al., Proc. Intl. Soc. Reson. Med., 13 (2005), 131 Cervical cancer diagnosis in vivo MRS + ex vivo (HR-MAS) normal control Invasive cervical carcinoma sagittal T2 TSE endovaginal coil 1.5 T TR 2 s TE 80 ms normal control 10 cm3 squamous cell carcinoma -CH2 normal uterine tissue Cho Cr squamous cell carcinoma uterine cervix Triglyceride CH2 1H MRS SV 3.4 cm3 PRESS spectra TR 1600 ms TE 135 ms biomarker of invasive cervical tumour (74% of tumours NOT in control p<0.001) 1H presat HR-MAS alipahtic region at 11.7 T 25C lipid and Choline levels markedly increased in squamous cell carcinoma biopsy M.M. Mahon et al. NMR Biomed. (2004), 17; 1 Pangenomic DNA microarrays eTUMOUR protocols GBM, OA, AII, MET, MEN. Not supervised analysis Hierarchical cluster Supervised LDA analysis MEN GBM MET A II OLIGO Lung Metastases vs Breast Metastases CPMG Gly Cho Cr Ala PCA Lung Metastases Breast Metastases Lac GBM high lipids content vs GBM lower lipid amount CPMG GBM with high lipids content GBM with lower lipid content and higher intensity for mobile metabolites PCA LipoMed, Inc. 3009 New Bern A venue Raleigh, NC 27610 www. lipoprof ile.com NMR LipoProfile LipoMe d, Inc. 0 4/99 P rod uced un der p atent license s to U.S. P aten t Nos. 4,93 3,844 a nd 5,34 3,389 LipoMed, Inc. 3009 New Bern A venue Raleigh, NC 27610 www. lipoprof ile.com NMR LipoProfile LipoM ed, Inc. 0 4/99 P ag e 2 of 2 P age 1 of 2 Pat ien t Na m e Pat ie nt Name Pa tien t ID Se x Age M 63 Bir th Dat e Spe cimen ID 7-29 -36 L M018 82 0 Dat e Co llect ed Date Re ceive d Dat e Re po rte d 8-24 -99 8-25 -99 8-2 6-99 Spe cim en ID Da te Rep or te d LM01 882 0 08-2 6-99 Physicia n N ame & Ad dr ess Phone: ( FA X: ( SUBCLASS LEVELS ) ) Comments VLDL Subclasses LDL Subclasses (mg/ dL Tr igly cer ide ) ( mg /d L C hole ste ro l) ( mg/d L Cho les ter ol) HDL Subclas ses (107) LIPOPROTEIN PANEL (21) (35) C oronary Heart Disease (C HD) Risk Categories (18) LDL Particles nmol/L Optimal* D es irable B orderline-High High Risk 1654 les s t han 1100 1100 - 1399 1400 - 1799 great er than 1800 (0 ) Pat tern A (large LD L) nm LDL Size Large HDL Large VLDL 19.1 22.0 - 20.6 20.5 - 19. 0 Low er-R isk Higher-Ri sk mg/ dL N egative R isk Fac tor Intermedi ate Posit ive Ris k Factor 16 greater than 40 40 - 21 less than 21 mg/ dL Lower-Ris k less than 7 Intermedi ate 7 - 27 Higher-R isk greatert han 27 35 (16) Pat tern B (small LDL) (5) (0) Lar ge Medium VLDL VLDL Sm al l VLDL (V5+ V6 ) (V1+V2) (V3+ V4) IDL Large LDL Medium LDL Sm all LDL ( L3) (L2) (L1 ) NMR LIPID PANEL RISK ASSESSMENT PANEL Elevated LDL Particles Atherogenic Dyslipidemia Total Chole sterol >1400 nmol/L 2 traits Smal l LD L Pattern B ( 20.5 n m) Re duce d Large HDL (< 21 mg/dL) Ele vated Large VLDL (> 27 mg/dL) Smal l HDL (H3+H4 +H5) (H1+H2) LDL Cholesterol D es ira b le 18 5 le ss th an 2 00 112 Triglyce rides O pt im al * le ss th an 1 00 Bo r de r line -H igh H ig h 20 0 - 2 3 9 2 40 o r g r ea te r De sir a ble Bo rd e rlin e- Hig h H igh R isk 1 00 - 1 29 1 30 - 15 9 g re at er th a n 1 6 0 Ne ga tiv e R isk F ac to r I nt er m ed ia te 6 0 or g re at er 5 9 - 35 m g/d L De sir a ble Bo rd e rlin e- Hig h 18 1 le ss tha n 2 00 m g/d L HDL Chole sterol Current NCEP Risk Categories m g/d L m g/d L Diagnostic Trait s of Atherogenic Dyslipidemia Large HDL - 37 Relative NMR frequency (Hz) Perfil metabólico Plasma P rod uced und er pa tent licenses to U. S. P aten t Nos. 4,933 ,844 an d 5,34 3,389 Lipoprotein diameter (nm) 20 0 - 4 0 0 P osi tive Ri sk Fa ct or les s t ha n 35 H igh 4 00 - 1 ,0 00 Determination of drug-toxicity (metabonomics) Rat urines after dosage of liver- and kidney-toxins 600 MHz-1H noesypr1d 64scans Sprague Dawley + Han-Wistar are standardised laboratory rat strains Hexachlorobutadiene Thioacetamid Hydrazin Sprague Dawley control Han Wistar control Determination of drug-toxicity (metabonomics) Coomans Plot showing 95% confidence level of classificlassification normal rat urines versus toxin treated urines Determination of drug-toxicity (metabonomics) Classification of hydrazine induced hepatotoxicity against renal cortical toxicity and controls based on rat urine 1D-NMR spectra Determination of drug-toxicity (metabonomics) Time course in rat urines after dosage of a-naphtylisothiocyanat (ANIT, liver toxin) 96-120h lactate 72-96 h acetate 48-72 h 32-48 h bile acids 24-32 h glucose 8-24 h 0-8 h control Metabolomic of gene alterations from DNA reparation 1D 1H spectra comparison of breast cell cultures with BRCA gene blocked and not blocked The most significant changes in the metabolite profile are marked Colaboration with Dra. Armengod FVIB Cell Culture differences CONTROL/AICAR Sample 124 Choline Glucose, amino acids betaine Lactate Lactate Creatine? Lysina? Asparagine? Glucose, amino acids Lípidos Colaboración Prof. JM. Esplugues F. Medicina Alanine Lípids Lípids Breast Cancer HR-MAS Visualización Apoptosis RMN a) Evolución temporal de 1H-ERM de un glioma transfectado HSV-tk tratado in vivo con glanciclovir b) RMI convencional de un glioma no tratado. c) Imagen de microscopía electrónica de transmisión (x5000) de células tumorales no tratadas d) Gotas de lípidos flechas vacías; cuerpos de apoptosis triángulos negros en células tumorales no tratadas e) En tumores tratados (4 días) gotas ricas en ácidos grasos poliinsaturados señaladas mediante flechas abiertas. Hakumäki y Brindle, Trends in Pharmac. Sci. ,24,3 (2003) Visualización Apoptosis RMN Imágenes de 1H RMI de un tumor de ratón tratado con glanciclovir posteriores a la inyección del agente de contraste C2-SPIO. (a) imagen previa a contraste; (b), (c), (d), (e) a 11, 47, 77 y 107 min después. Las áreas de apoptósis, captan contraste, áreas de pérdida de intensidad (flecha blanca). Imágenes inferiores sustracciones. Hakumäki y Brindle, Trends in Pharmac. Sci. ,24,3 (2003 CLAS clasificación module Metabonomic of Folicular Fluid Proteins vs Metabolites (0.1-3KDa) Contribución Clínica y Biomédica RMN Alto Campo • • • • Diagnóstico Clínico Biofluidos Diagnóstico Clínico Tejidos Estudios Biomédicos Líneas Celulares Estudios Farmacocinéticos: Tejidos Biofluidos • Desarrollo Biomarcadores: Diagnóstico Diseño Fármacos • Mejora Compresión Bioquímica Patologías NMR Applications 1H( MHz) Magnetic Field Intensity (T) Size (cm) 9,6 - 100 0,25 - 3,0 < 100 d5 MRI + 41 - 300 MRS (biomedicine) 1,0 - 7,1 < 30 d2 Miniimaging 300 - 400 7,1 - 9,4 < 12 # 0,005 mouses NMR Microscopy 200 - 600 4,7 - 14,1 < 2,5 0,00005 insects, materials, embrions 9,4 - 21 < 40 Å Type resonance frequency MRI + MRS (clínical) NMR 400 - 900 multidimensional B.Celda Spatial resolution (mm3) Application System human body small animals (rabits, mouses…) proteíns, DNA, tissues ... FIHCV-06 Aknowledgments Hospital La Ribera-Alzira Dr. J. Piquer Dr. E. Mollá Dr. A. Revert Dr. P. Ferrer IVO- Valencia Dr. J. Cervera Dr. A. Marhuenda Dr. A Menendez F. Medicina (UVEG)-Valencia Prof. F. Pallardó Prof. J.M. Rodrigo Prof. M. Cerdá Dr. M. Mata Hospital Clínico-Valencia Dr. J. Del Olmo Dr. J. Talamantes Fundación Investigación H.Clínico Clínica Quirón Dr. L. Martí-Bonmatí Bruker Biospin Lab of Applications Dr. M. Piotto Dr. O. Assemat B.Celda eTUMOUR Consortium Prof. C. Arús (UAB-Barcelona) Dr.F. Howe (SGHMS-London) Prof. A. Heerschap (UMCN-Nijmengen) Prof. L. Buydens (KUN-Nijmengen) Dr. C. Segerbart (INSERM-Grenoble) Mr. M Lluch (Microart-Barcelona) Dr. A. Capdevilla (HSJD-Barcelona) Dr. G. D’Incerti (PQE-Milan) Dr. L. Visani (PQE-Milan) Prof. S. Van Huffel (KUL-Leuven) Dr. P. Kreisler (Siemens-Etlinger) Dr. A. Klaasen (SCITO-Paris) Dr. M. Robles (UPVLC-Valencia) Dr. S. Wolfhard (DKFZ-Heidelberg) Dr. C. Brevard (Bruker-Biospin-Wissembourg) Dr. R. Grundy (BU-Birmimghan) Dr. F. Berger (INSERM-Grenoble) Dr. J. Calvar (FLENI-Buenos Aires) Dr. W. Gajewicz (MUL-Lodz) UVEG-Valencia Dr. V. Esteve; MB Martínez-Granados E. Piñero; R. Ferre IIIGERMN